
In this blog, we’ll walk you through how a scientist at Addgene uses Snapgene to confirm the sequence of a plasmid and we’ll highlight some of the new features available on our website through our Snapgene powered maps and sequence analysis tools. We regularly use Snapgene for our quality control process because of its expansive feature library and useful tools. We’ve also talked about how our new Snapgene generated maps provide improved feature detection with an easy to use interface. This will determine whether or not it will be read correctly.We’ve talked a lot about the quality control process at Addgene by introducing our new sequencing partner seqWell and going into detail about how we use next generation sequencing results to perform quality control on deposited plasmids. In addition, the combination of restriction enzymes you use determines how a genetic sequence will fit in a construct. This is of particular importance because you don’t want to use an enzyme that can cut somewhere in the middle of a genetic sequence. There are a number of things to consider when applying restriction cloning, such as which restriction enzymes to use. SnapGene allows you to simulate this process either by using the protocols it has available or creating your own. This process is performed in the lab following a given protocol. A number of cut sequences can be joined together with a process known as ligation. Restriction cloning is a method of editing genetic sequences by cutting them with restriction enzyme at suitable restriction sites. The Sequence view also shows the features seen in the map view, and adds an extra feature, a red mark that indicates the stop codons of your sequence. The Sequence view shows the characters in the sequence is convenient when editing sequences because it allows you to pinpoint the critical locations in the sequence. This information is important for estimating the cost of synthesizing your sequence and is useful in a number of calculations. This includes the number of base pairs, the number of amino acids and the molecular weight of the sequence in Daltons. The Features view, which is accessed by clicking on Views and selecting Features, shows some information about the sequences you have. These views allow you to see features included in your file, the restriction enzymes in your sequence, the actual characters in the sequence, and your primers. Other views can be accessed by selecting one of the options in the Views menu. If the length and direction of the reading frames feature are the same as the length and direction of your gene’s feature, the gene will likely be transcribed correctly. These features allow you to see the reading frames of the sequences you have in your file, and highlights the parts of your construct that hold given sequences.Ĭlicking on show translate on the left adds some features that show the reading frames of each sequence.
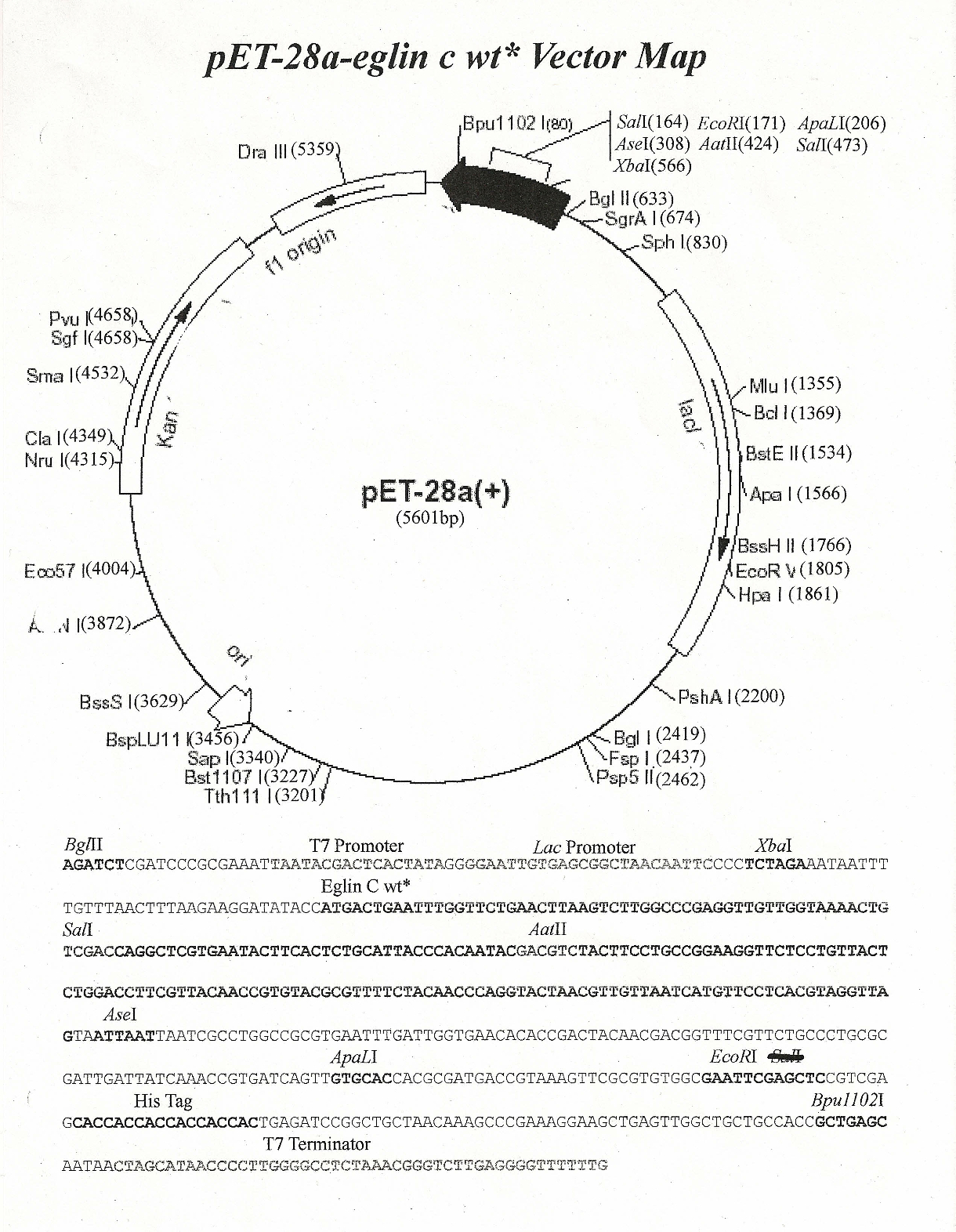
The default view is the map which hides the letters in the genetic sequence to create space for visual features. There are a number of ways you can view a genetic sequence in SnapGene. This is extremely useful in a field like synthetic biology where existing sequences are combined to form new ones that allow organisms to perform novel functions.
#Snapgene viewer restriction enzyme digest software
SnapGene is software that allows you to visualize and edit genetic sequences.


 0 kommentar(er)
0 kommentar(er)
